What are viruses made of?
Given their daunting biological task (infecting a host) and their limited biological capital, viruses have no choice but to be thrifty. While every strain of virus has its own unique size and shape, the primary function of a virus’s biological “stuff” is pretty standard: transmit a copy of their genetic material from an infected cell to an uninfected cell. What is this biological stuff, anyway? Let’s take a look!
The Building Blocks of Viruses
It is hard for scientists to know exactly when viruses first emerged, but we do know that viruses originated at least as early as the first cells, around 4 billion years ago. Some even think viruses showed up before there was cellular life. Either way, viruses have had a really long time to get it right. As it turns out, they are efficient molecular systems with a metastable structure (more on this below!). When you get down to brass tacks, the basic functions of a virus are to protect its nucleic acid genome, and be able to attach and enter a host cell. Here, we will discuss the building blocks of viruses that help them do this job.
The Viral Capsid
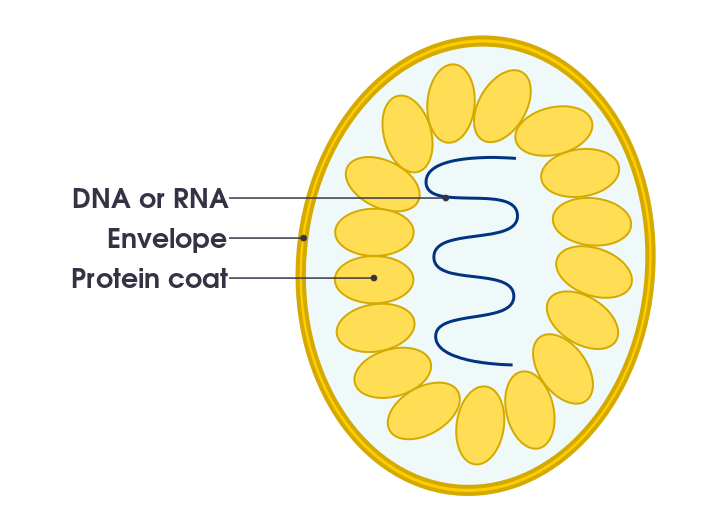
A simple sketch of a virus: nucleic acid genome, surrounded by a protein coat (capsid), additionally surrounded by a membrane (envelope)
There are all sorts of virus shapes and sizes. However, all virus particles have a protein coat that surrounds and protects a nucleic acid genome. This protein coat is called a capsid, and the instructions for making the protein subunits of the capsid are encoded in the nucleic acid genome of the virus. The structure resulting from the combination of the capsid and the nucleic acid genome is called the nucleocapsid. A single, fully assembled, infectious virus particle is called a virion.
Viruses strive to be as simple as possible while still maintaining their basic function, a concept that scientists call genetic economy. This means that the nucleic acid genome of the virus can be very tiny, providing instructions for only a few types of capsid protein subunits, each of which get produced in large numbers. Amazingly, these protein subunits can self-assemble in a stable and repetitive way, with each subunit forming the maximum number of contacts with the next subunit. As a result, the capsid of viruses is like a coat of armor for the nucleic acid genome, and the repetitive nature of the identical protein subunits give nearly all viruses geometrical symmetry.
Yet, the viral capsid can’t be so stable that it’s nucleic acid genome cannot escape into the host cell. As mentioned above, the capsid of a virion is metastable — with the right kind and amount of perturbation, the capsid can become undone, allowing host cellular machinery to get access to the viral genome. The ability of the virion to disassemble is afforded by the fact that viral capsid subunits are NOT covalently bound, and will release from each other with the appropriate signal.
Some viruses, such as the now famous coronavirus, also have a lipid membrane that surrounds the capsid. Studies on viruses indicate that this lipid membrane is “stolen” from host cells that the virus infects. Moreover, the lipid membrane of a virus contains specific viral proteins that are “tagged” with carbohydrates (sugars). These sugar-protein complexes are found on the surface of a virus particle, and are called glycoproteins. While glycoproteins are not specific to viruses (there are many examples of glycoproteins throughout all life), they do provide a way for viruses to attach themselves to host cells. Since viral glycoproteins are one of the key ways viruses can infect cells, many scientists are working on medicines that can impact how the glycoproteins work in order to prevent viral illnesses in people, pets, and plants.
The Viral Nucleic Acid Genome
In addition to being varied in their shapes and sizes, viruses also demonstrate diversity when it comes to their nucleic acid genomes. The primary function of a viral genome is to store the instructions for building more virus particles. Depending on the type of virus, the size of a viral genome can range from ~2 kilobases to 2 megabases — that is a range between 2,000 and 2,000,000 of nucleic acid “letters” in length — depending on the complexity of the virus structure.
Many virologists classify viruses based on what kind of nuclear genome they have, which can be made out of DNA or RNA, exist as a single strand or a double strand, and can be either circular, linear, or segmented (for more in depth descriptions of this, see this blogpost on the “Baltimore Classification” system by virology professor, Vincent Racaniello). Regardless of which type of genome a virus has, there are two main routes for packing it: viruses can either assemble their capsid shell around their nuclear genome, or viruses can make a capsid shell, and insert their nuclear genome into it. Viruses also need to make sure that they are packaging their genomes, and not the genomes of their host cells. This is often accomplished through a special signal sequence inside the viral genome that can bind to the viral capsid, helping to ensure that the “right” nucleic acids are included in each virion.
Because there are millions of different viruses, there are millions of different viral genomes. So far, scientists have mapped the genomes of 75,000 viruses, but that is merely a fraction of what is out there. As next generation sequencing and analysis continues to grow in its sophistication, scientists will continue building knowledge when it comes to viral genomes!
References
Gelderblom, H. R., (1996). Structure and classification of viruses. In S. Baron (Ed.), Medical microbiology (4th ed.). University of Texas Medical Branch.
Holmes, E. C. (2011). What does virus evolution tell us about virus origins? Journal of Virology, 85, 5247-5251. doi: 10.1128/JVI.02203-10
Knipe, D. M., & Howley, P. M. (Eds.). (2013). Fields virology. Lippincott Williams & Wilkins.
Woolhouse, M., Scott, F., Hudson, Z., Howey, R., & Chase-Topping, M. (2012). Human viruses: discovery and emergence. Philos Trans R Soc Lond B Biol Sci, 367, 2864–2871. doi: 10.1098/rstb.2011.0354