How do scientists "see" viruses? Part 2: qPCR
qPCR—quantitative PCR (polymerase chain reaction), sometimes also called real-time PCR—is a method for detecting nucleotide sequences (in this case, signatures of viral genomes) through a quick and often quantitative method.
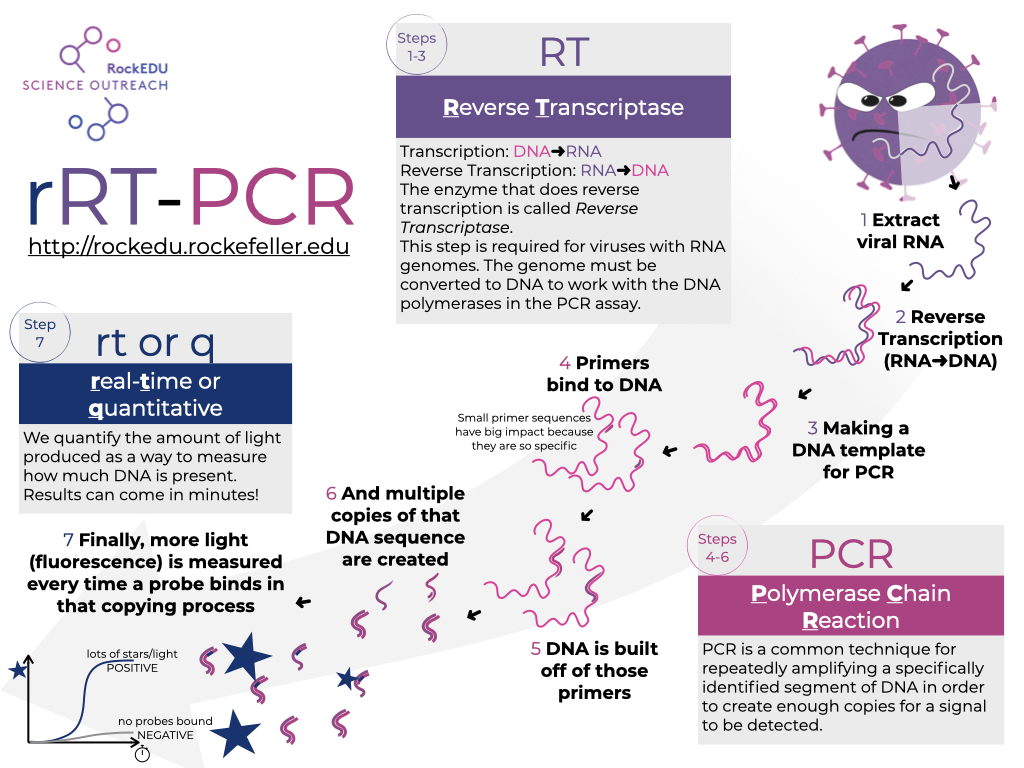
We can simulate the detection of a virus by this method using many of the components scientists have used to design and implement tests for SARS-CoV2 since its discovery in December 2019. Viruses like SARS-CoV2 with genomes stored as RNA, the test has an added step for reverse-transcription of the RNA to DNA and these tests are then often referred to as RT-PCR. The entire process is summarized in the following infographic, which is also available for download in the Save & Share menu.
In the subsequent steps of the activity, we will 1) explore a key component of the specificity of PCR in general, 2) model the RT-PCR test for SARS-CoV2 in silico, and 3) model the added component that can make these tests quantitative and reasonably fast, making them a gold standard for virus identification in an infection.
PCR Practice Tutorial
We will model the function of the test by analyzing how the PCR step of the RT-PCR works in detail. So, let’s get familiar with the online platform and the PCR test itself through this short preview tutorial. PCR works such that DNA sequences added in the test reaction will only bind to complementary DNA if it is present in the patient sample. These specific sequences are called primers and are designed specifically for the polymerase enzyme (aka the make-more-DNA-polymer machine) to make more DNA off of this as a starting sequence. With two primers that initiate DNA synthesis in opposite directions, several cycles of this reaction will produce many copies of that DNA segment—eventually enough to be detected.
Let’s give this a try by using the online genome database to search for all possible matches by trying the primers that we use in our PCR LAB Experience where we look for human mitochondrial DNA that is the same between all humans:
Fwd: TTAACTCCACCATTAGCACC
Rev: GAGGATGGTGGTCAAGGGAC
Note that when we run this experiment in the lab, it’s done as a standard PCR (amplifying DNA and reading out the presence or absence of a sequence by the presence or absence of a band in an agarose gel) but the read out from our primer-binding search in silico (aka on a computer) will give us the same kind of information here.
The downloadable step-by-step tutorial will guide you through using Primer-BLAST and making sense of the outputs. You can find the tutorial PDF in the Save & Share menu.
In this sample name, where does it tell you this is from human mitochondrial DNA?
How long is this PCR product?
Are you surprised by how many sequences matched? Why or why not?
RT-PCR to find SARS-CoV2
For the RT-PCR test to give a positive read-out (aka you have SARS-CoV2 in your body), a few things have to happen. So, let’s go through each one:
A sample is swabbed from inside a patient’s nose or on the back of their throat. This sample should pick up some of the patient’s cells. {If you have come to visit our PCR LAB Experience, then you have done just this by using a toothpick to scrape off some cheek cells.} Once the sample is collected, it must be prepared by adding reagents to lyse—or break open—the cells.
What types of molecules and things would you expect to be in this broken-open-cell-soup? Make a list or draw a picture:
Some of this broken-open-cell-soup (lysate) is mixed with the pieces needed for the RT-PCR reaction and it is put into a machine.
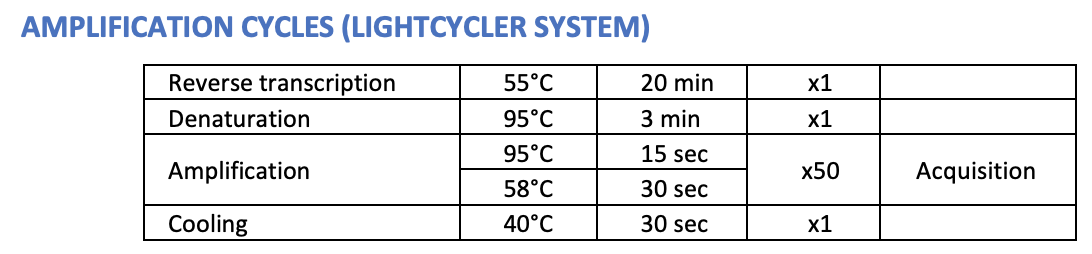
What is happening for the first 20 minutes (the first step)? What kind of molecule is being made?
The RT-PCR test can specifically look for SARS-CoV2 by using specific sequences of DNA that will only bind to complementary DNA in the patient sample. These specific sequences are called primers and are designed specifically for the polymerase enzyme (aka the make-more-DNA-polymer machine) to make more DNA off of this as a starting sequence. With two primers that initiate DNA synthesis in opposite directions, several cycles of this reaction will produce many copies of that DNA segment—eventually enough to be detected.
In the steps below, you can go through your own simulation of how the test works using online datasets of genome sequences and an algorithm for searching them known as BLAST. We will specifically use Primer-BLAST to look for the genomes that will be detected by our chosen primer sets.
Let’s look for the 1st round SARS-CoV2 test, which looks for a sequence of DNA that will make the viral E protein (learn more about this and the other parts of coronaviruses in this great explainer video by the SimpleBiologist)
Fwd: ACAGGTACGTTAATAGTTAATAGCGT
Rev: ATATTGCAGCAGTACGCACACA
Follow the steps on the COVID-19 Tutorial page available as a PDF in the Save & Share menu
Next, let’s look for the sequence currently most definitive for SARS-CoV2
Fwd: GTGARATGGTCATGTGTGGCGG
Rev: CARATGTTAAASACACTATTAGCATA
You can follow the COVID-19 Tutorial again or try to do it yourself
Were the matching sequences uniformly named? Why or why not? Why is this important to notice and reflect on as a scientist?
quantitative/real-time PCR
The final step is the detection or read-out of the assay. qPCR is interesting in that it uses a relatively sophisticated fluorescence technique that can produce a fluorescent signal if the specific DNA sequence is being transcribed, giving a read-out in real-time, hence why it is often called real-time PCR or quantitative PCR.
When running this test, both the forward and reverse primers (as tested above) are included, as well as a probe sequence with specificity to the transcribed sequence. While the Primer-BLAST platform doesn’t specifically allow for adding such a third sequence, you can approximate it by replacing the forward or reverse primer with the probe sequence.
One will work well and the other will not…can you figure out which one and why?
Consult the WHO protocol for specific probe sequences. This is currently an open-ended exploration. More step-by-step guidance will be added here soon.
References
https://www.newyorker.com/news/news-desk/what-went-wrong-with-coronavirus-testing-in-the-us
https://www.theatlantic.com/health/archive/2020/03/how-will-coronavirus-end/608719/