Coronavirus Biology
There are many types of coronaviruses. In fact, scientists have been studying coronaviruses since the 1960s. But, it was not until 2020 that “coronavirus” became a household name due to the global pandemic caused by a novel coronavirus species, SARS-CoV-2. What is a coronavirus? How does a coronavirus infect cells? Why are coronavirus genomes so large? Let’s explore.
What is a coronavirus?
First recognized by scientists in 1968, coronaviruses (CoVs) are among the largest family of viruses currently known. These infectious particles were named based on their “spikey” appearance. At the time of their identification, scientists had only recently been introduced to the electron microscope. Upon getting their first views of CoV morphology, scientists noted that the club-shaped spikes emanating from the surface of CoV particles looked like images of the sun shared by NASA. More specifically, the scientists thought that CoVs resembled the coronal mass ejections that appear during a solar storm. Hence, this is where coronaviruses get their name. In Latin, corona means crown. 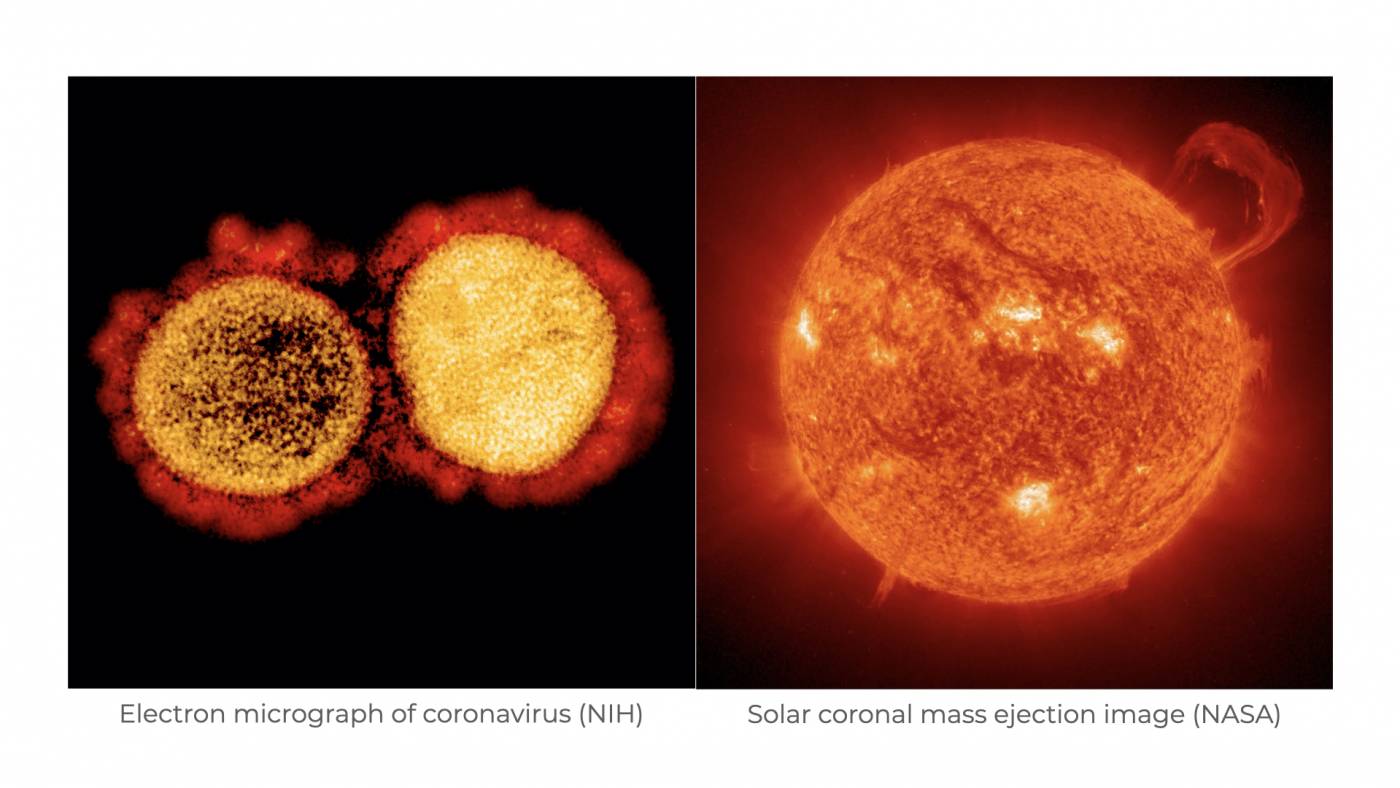
While there are many different species of CoV that infect different hosts, there are a few core features that exist among all CoVs. First, all CoVs contain very large, positive-sense RNA genomes that are enclosed within a helical nucleocapsid. These nucleocapsids are surrounded by a lipid membrane that is “stolen” from the host cell. Interestingly, while the average size of RNA virus genomes tends to hover around 9 kilobases (that is 9,000 nucleotide “letters” in length), the genome size of CoVs is closer to 30 kilobases — just over 3 times the average. In virology speak, CoV genomes are HUGE.
How is the CoV genome organized?
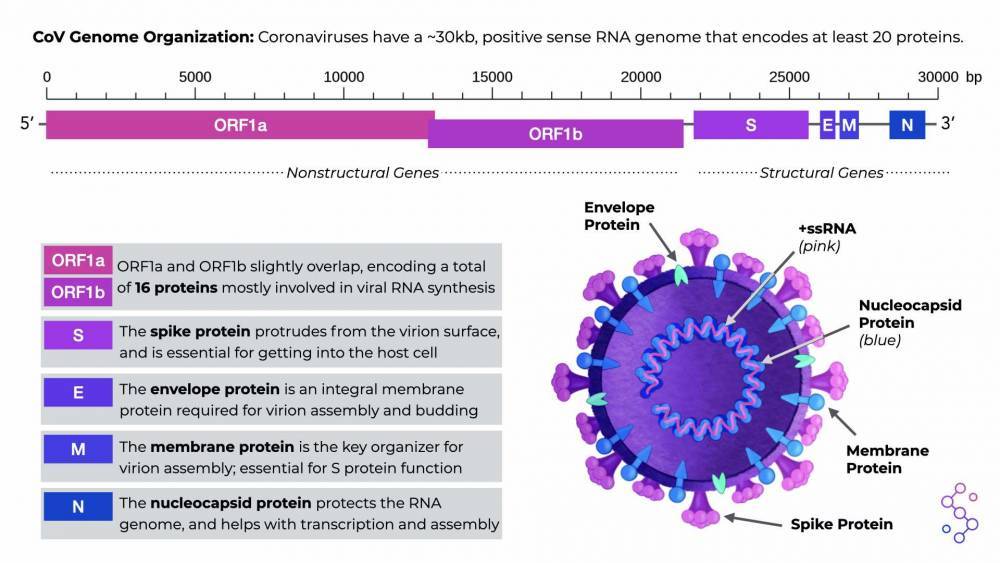
There are several sections of the CoV genome, known as open reading frames (ORFs), that serve as the instructions for making CoV proteins. All CoVs have two, really long ORFs that are translated into equally long “polyproteins,” which are then cut up to make 16 individual, non-structural proteins. These nonstructural proteins are responsible for basic but highly important functions that mostly involve viral RNA synthesis. All CoVs also have an additional 4 ORFs for making the structural proteins of the virus: spike (S), envelope (E), membrane (M), and nucleoprotein (N). There are other proteins that CoV genomes can encode, but this can differ depending on the specific CoV species.
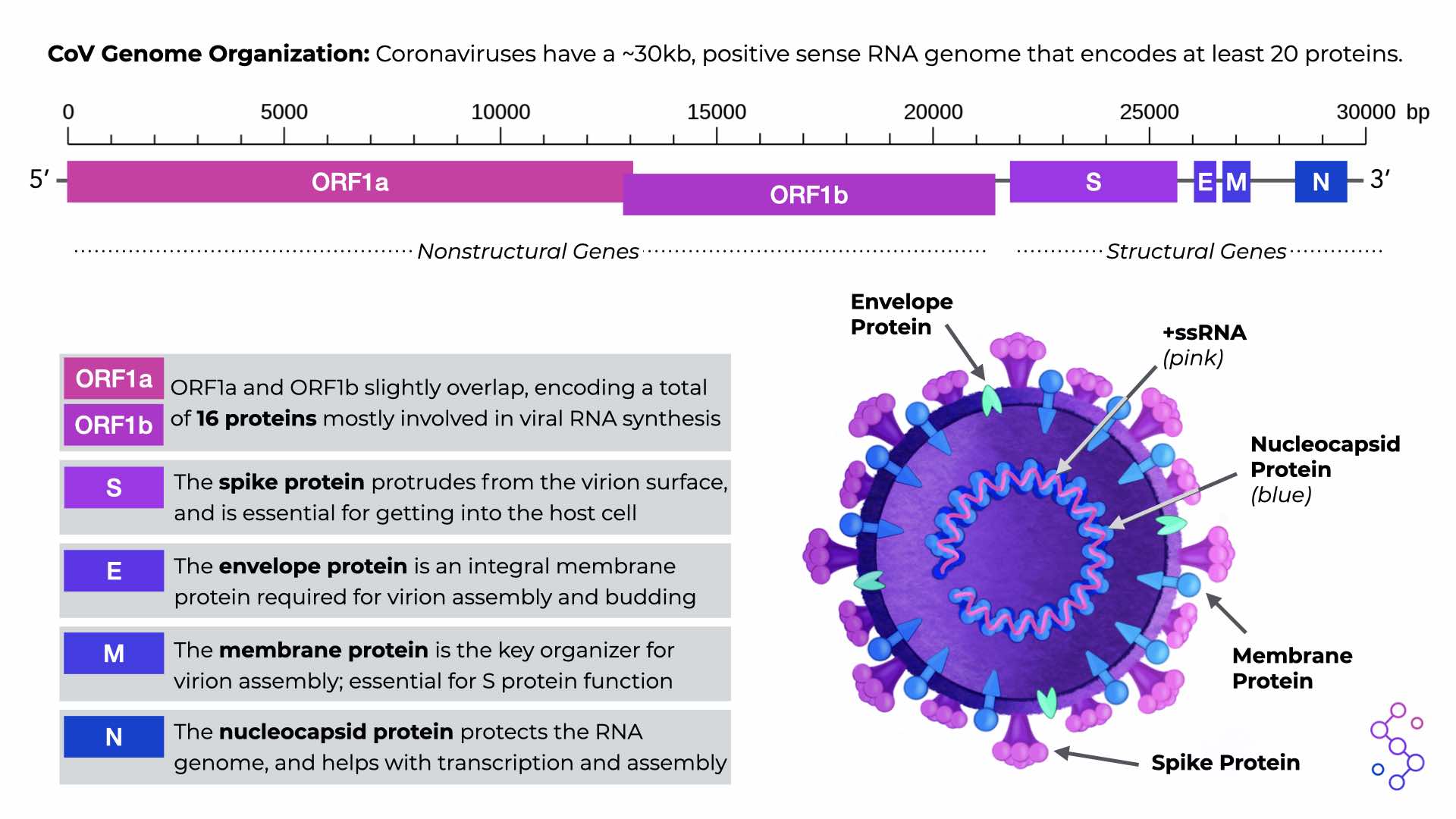
CoV Genome Organization. Download the PDF version in the “Save & Share” tab.
How did the CoV genome get so big?
RNA viruses tend to have low replication fidelity.This means that there are more chances for errors to be incorporated into their genome with every round of replication. But, these RNA virus mutations can be a good thing — for viruses, at least! Mutations are the currency for natural selection, and a high mutation rate in RNA viruses can help select for populations of viruses that are more infectious. However, too many mutations in a viral population can impact overall function, and render a virus species noninfectious. Virologists believe that the small size of most RNA genomes is how these entities balance their high mutation rates with the survival of their species.
Yet, CoV genomes are three times larger than the average RNA virus genome. Why? While we can never gain access to the precise set of steps that took place over the course of CoVs evolutionary history, we can look at various factors and genomic information as forensic evidence. Interestingly, CoVs have a key feature that other, smaller RNA genome viruses do not have: a molecular eraser called ExoN (encoded by the nonstructural region of the CoV genome). While the precise mechanism of ExoN action is still under scrutiny, scientists believe that ExoN helps prevent and/or repair mutations during genome replication.
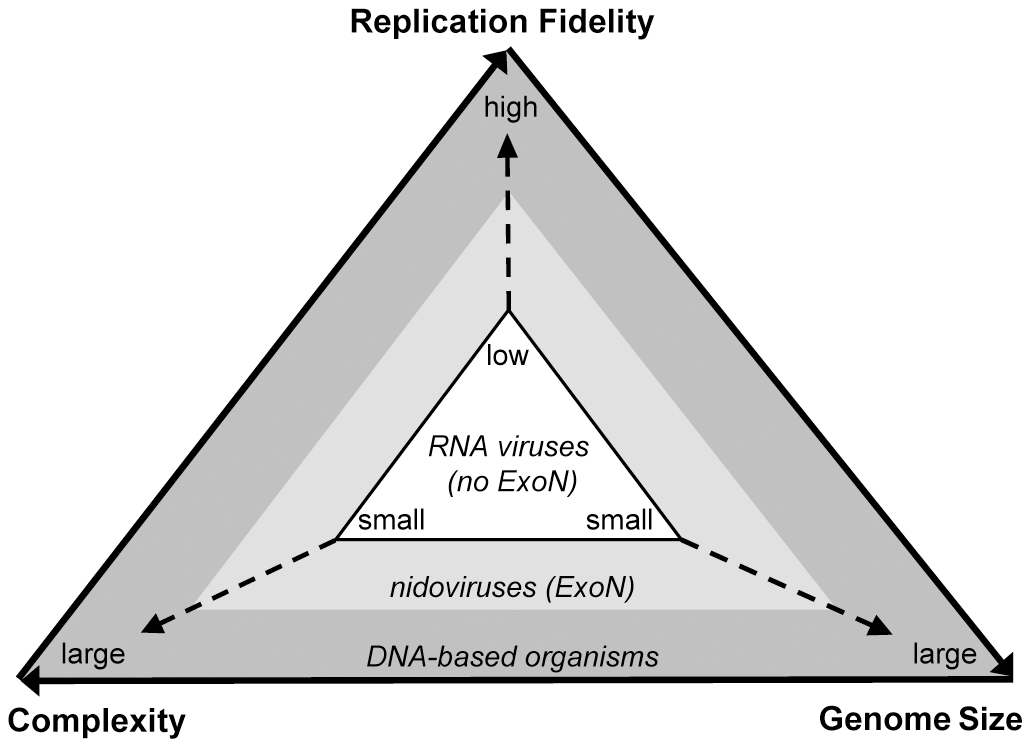
A schematic illustrating the relationship between replication fidelity, genome size, and complexity (PLoS Pathog 7(9): e1002215).
To answer the question of “how did CoV genomes get to be so big?” we need to look at the evolutionary history of CoVs in the context of ExoN. Scientists believe that viruses belonging to the nidovirus order (containing the CoV family of viruses) acquired ExoN from a common ancestor. This resulted in increased genome replication fidelity and allowed CoVs to increase the size of their genome over time. Essentially, the acquisition of ExoN function helped CoVs strike a balance between a suitable mutation rate, and genome complexity.
How does a CoV get into a host cell?
CoVs use their spike proteins as a key to get into the host cell. More specifically, the spike protein will bind to a specific receptor on the surface of a host cell, which initiates a cascade of reactions that causes the host cell membrane to surround the virion, and bring it into the cell via endocytosis. The virion then sits in a vesicle within the host cell, and normal cellular processes lead to a change in the pH inside the vesicle (it becomes more acidic). The acidic environment causes the virion to disassemble, and it releases its viral genome into the cell’s cytoplasm. Here, the CoV genome is acted upon by host cell machinery, and the building blocks for making new virions are made and packaged into additional cellular vesicles, where they self assemble. Finally, the newly made virions leave the cell through exocytosis, and are able to infect new cells.
References
Clinton Smith, E., Sexton, N. R., & Denison, M. R. (2014). Thinking outside the triangle: Replication fidelity of the largest RNA viruses. Annual Review of Virology, 1, 111-132. https://doi.org/10.1146/annurev-virology-031413-085507
de Haan, C. A., Smeets, M., Vernooij, F., Vennema, H., & Rottier, P. J. (1999). Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. Journal of virology, 73, 7441–7452.
Fehr, A. R., & Perlman, S. (2015). Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol, 1282, 1–23. doi:10.1007/978-1-4939-2438-7_1
Li, G., Fan, Y., Lai, Y., Han, T., Li, Z., Zhou, P., Pan, P., Wang, W., Hu, D., Liu, X., Zhang, Q., & Wu, J. (2020). Coronavirus infections and immune responses. Journal of Medical Virology, 92, 424, 432. https://doi.org/10.1002/jmv.25685C
McBride, R., van Zyl, M., & Fielding, B. C. (2014). The coronavirus nucleocapsid is a multifunctional protein. Viruses, 6, 2991–3018. doi:10.3390/v6082991
Nakagawa, K., Lokugamage, K. G., & Makino, S. (2016). Viral and Cellular mRNA Translation in Coronavirus-Infected Cells. Adv Virus Res, 96, 165–192. doi:10.1016/bs.aivir.2016.08.001
Nga, P. T., Parquet, M., Lauber, C., Parida, M., Nabeshima, T., Yu, F., Thuy, N. T., Inoue, S., Ito, T., Okamoto, K., Ichinose, A., Snijder, E. J., Morita, K., & Gorbalenya, A. E. (2011). Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS pathogens, 7, e1002215. https://doi.org/10.1371/journal.ppat.1002215
Virology: Coronaviruses. (1968). Nature, 220, 650. https://doi.org/10.1038/220650b0