Symbiotic evolution with microbes
Symbioses are the ultimate examples of success through collaboration and the powerful benefits of intimate relationships. — David Relman
In biology, there is a tendency to walk a reductionist path, breaking every system down to individual components in order to understand how a biological system works. This strategy has certainly shed light on many fundamental principles pertaining to life on earth, leading to immeasurably impactful discoveries. However, as new technologies emerge and perspectives widen, we are able to take a more holistic viewpoint in our quest to understand the underlying complexities of life.
This is particularly true in the context of evolution. The study of evolution has historically centered on how competition at the individual level — i.e. “survival of the fittest” — ultimately impacts the phenotypic frequencies in a population. This notion of natural selection, which was independently developed by both Charles Darwin and Alfred Wallace (jointly published in 1858), established the idea of speciation from a common ancestor. Around the same time, Gregor Mendel, a monk from the Austrian Empire (now Czech Republic), developed the underpinnings of modern genetics through pea plant hybridization experiments.

Charles Darwin (left) and Alfred Wallace (right). Source: Popular Science Monthly Volume 73 [Public domain], via Wikimedia Commons

Evolutionary theorist and biologist, Lynn Margulis. Source: SINC [CC BY 1.0], via Wikimedia Commons
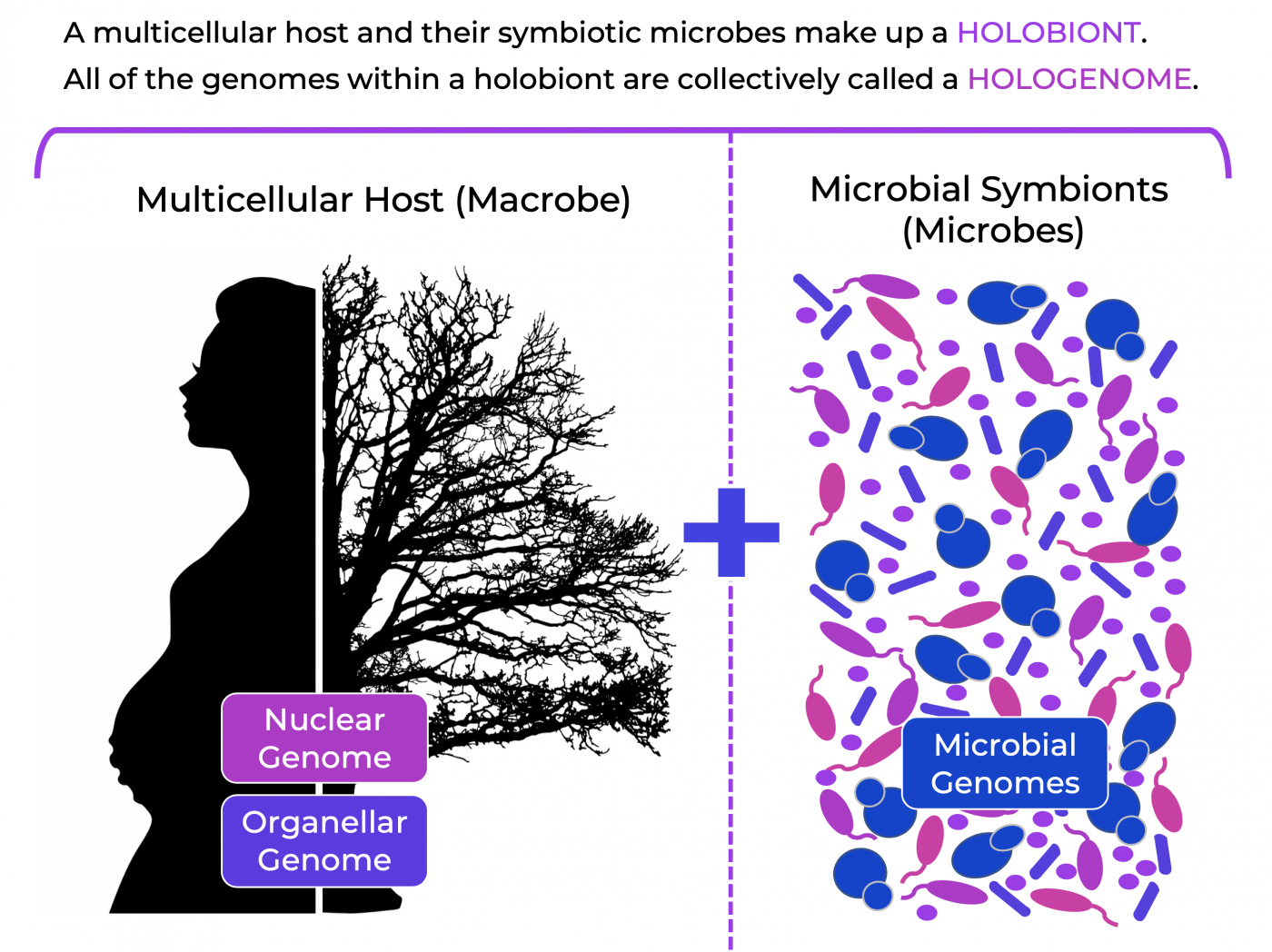
If “units of life” consist of a group of organisms living in a symbiotic relationship (holobiont), and that within this exists a network of genomes (hologenome), then it is important to consider whether this represents a unit of selection during evolution (i.e. do microbial symbionts play a role in the evolution of higher organisms?). Expanding on our current knowledge of evolution, the Hologenome Theory of Evolution posits that microbial symbionts significantly influence how a host organism responds to evolutionary pressures (5). Given this expanded view, natural selection might actually be a function of genetic variation among hologenomes, which allow the holobiont to adapt to environmental changes.
Symbiosis and Natural Selection of the Hologenome
Symbiosis, or, in biological terms, the relationship between two or more organisms, can be classified in several ways:
- Commensalism describes the relationship between symbionts where one receives a benefit, whereas the other receives no benefit or harm (neutral)
- Mutualism occurs when a benefit is experienced by all symbionts
- Parasitism is when one symbiont benefits at the expense of another symbiont
Microbial parasitism has been a huge focus throughout modern scientific history, namely due to the importance of understanding human infectious disease (i.e. germ theory), the impact that pathogens have on the agricultural industry, or other factors of economic importance. But “infection” is not always a bad thing, and studies on all types of symbiotic relationships are shedding light on the ecosystems within a holobiont. Using human hosts as an example, we know that different regions of the body serve as distinct ecological niches that house a defined set of microbes (i.e. your belly button is a different ecological niche compared to your eye lids or colon). In general, symbiosis between host and microbe can vary in the following ways (6):
- Host-microbial specificity
Different microbes will favor colonization of different hosts over others, or will even display preference for ecological niches within a host. - Symbiont acquisition and maintenance
While we don’t truly understand the mechanisms underlying the acquisition and maintenance of microbial symbionts, these factors clearly impact the status of the holobiont. In general, hosts acquire symbionts through a) horizontal transfer from the environment, such as through food intake; b) vertical inheritance from parent, such as through a vaginal birth delivery; or a combination of a) and b). - Functional output of symbiotic relationship
Symbiotic microbes can carry out many functions required for the health of the holobiont, such as synthesis of essential amino acids (7), or the production of metabolites (8). Symbionts can be obligate (required for survival) or facultative (optional). - Response of host to “infection”
Will the symbiont set off an immune response in the host, or will it assimilate into the environment afforded by the host?
All of these factors can create sources of variation within the hologenome, which can be acted upon by natural selection, leading to the “survival of the fittest” holobionts. This ultimately impacts the hologenomic frequencies in a population. However, in the absence of selective pressure, variation at the holobiont-species level can also arise due to functional redundancy. This explains why some hosts of the same species have a core set of microbiota at higher taxonomic levels (i.e. order or phylum), but not at the microbe-species level. For example, there are many different species of bacteria that produce lactic acid as a fermentation product, but that does not change how lactic acid functions — this means that lactic acid bacteria are taxonomically diverse at the species level, but when considered at the functional level, they are all very similar.
While we are just beginning to uncover the incredible complexity of host-microbe symbiosis and coevolution in the holobiont, we do know that the host genome (GH) interacts with the genome of microbial symbionts (GM), and each genome within the hologenome interacts with the environment (E).
Taken together, evolution can be viewed in terms of the holobiont: GH x GM x E
Limitations of the Hologenome Theory of Evolution
The hologenome theory of evolution, which expands on the current Darwinian model, is not without limitations and refutations. For instance, arguments are being made that a holobiont is a community as opposed to an individual evolutionary unit, and therefore cannot be acted upon by natural selection (9). But say that holobionts are indeed individual units that are selected upon, are all genomes within a hologenome inherited to the same degree, and if so, are they subject to equivalent evolutionary pressure? We already touched on the idea that the microbiome can be acquired through vertical as well as horizontal transmission. But evidence is showing that a horizontally-acquired microbiome in a host can in fact be passed down to the progeny of said host, echoing traces of Lamarckian evolutionary theory on the “inheritance of acquired characteristics” (4).
Although Jean-Baptiste Lamarck, a 19th century French naturalist, did not consider the microbial world when crafting his evolutionary theory, it is interesting how his principles apply to the evolution of the holobiont. However, until we can determine the degree to which horizontally-transmitted microbiomes are maintained in populations, much will remain unanswered.
Fundamentally speaking, the evolution is characterized by changes in genetic frequency in a population over successive generations. Whether this applies to individual genomes or a hologenome remains to be answered. We are just beginning to build knowledge around the hologenome concept, and as research advances, we will start filling in the gaps in our current understanding.
Holobionts: Beyond Humans as Hosts
The human microbiome is a hot topic, both in science research and in popular media. More and more, we are learning about the impact of the microbiome on human health and well-being. But, these studies represent a very anthropocentric understanding of symbiosis between a host and it’s microbial symbionts. While it is valuable for us to know and understand such teachings, we must acknowledge that the evolution of nearly every eukaryotic organism on this planet has occurred in the context of microbial symbiosis.
One of the earliest known example of microbial symbiosis dates back approximately 1.5 billion years when an archaebacterial cell was infected with a smaller proteobacterial cell. Over time, this proteobacterial cell became part of the necessary infrastructure within the archaebacterial cell, and became mitochondria. This incredible milestone in earth’s natural history represents Lynn Margulis’s endosymbiotic theory of eukaryotic origin, or how bacterial symbiosis led to the formation of the first eukaryote (10).
Another extraordinary evolutionary milestone resulting from symbiosis occurred approximately 460 – 700 million years ago, when the first “plants” colonized land. More specifically, scientists believe that an ancient species of fungus, which had the ability to grow root-like filamentous structures, or mycorrhizae, assisted a phototrophic algae as it transitioned from water to land (11, 12). The evolutionary predecessors of this ancient fungus, which belong to the glomeromycota division of the fungal kingdom, live almost exclusively as symbionts with current land plants.
Symbiotic relationships may not always represent a favorable situation for a host cell. Indeed, infection with a parasitic symbiont could have a deleterious impact on a species. However, parasitic symbionts also push their hosts to resist their own demise, and put up a cellular defense. As any living cell can be considered a host, it is important to recognize that both prokaryotes and eukaryotes have developed some type of immune system, the evolution of which was influenced, at least in part, by parasitic symbionts (13). One of the most valuable discoveries in biology today — the CRISPR/Cas9 genome editing system (14) — harnesses the technology of the prokaryotic immune system, and will undoubtedly revolutionize the face of what is possible in biology.
Humans have been working to understand the mechanisms underlying evolution for less than 200 years. As the scientific toolkit grows in its sophistication, so does our understanding of biological processes. However, new insights also provide clues into how much there is yet to discover. With next generation sequencing technology, we are able to take a more comprehensive view on the nature of symbiosis, and begin to shift the evolutionary paradigm to include the incredible role of the microbial world.
Bibliography
- Behjati S, Tarpey PS. What is next generation sequencing?. Arch Dis Child Educ Pract Ed. 2013;98(6):236-8. [PDF]
- Moran NA. Symbiosis. Current Biology. Volume 16, Issue 20, 2006, pp R866-R871. [PDF]
- “Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis.” Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis, by Lynn Margulis and Fester René, MIT Press, 1991.
- Bordenstein, Theis, Waldor. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLOS Biology. 2015;13(8). [PDF]
- Zilber-Rosenberg I and Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiology Reviews, Volume 32, Issue 5, 1 August 2008, Pages 723–735 [PDF]
- Webster NS. Cooperation, communication, and co-evolution: grand challenges in microbial symbiosis research. Frontiers in Microbiology. 2014;5. [PDF]
- Wilson, ACC, Ashton PD, Calevro F, Charles H, Colella S, Febvay G, et al. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Molecular Biology. 2010;19:249–258. [PDF]
- Nicholson, Holmes, Kinross, Burcelin, Gibson, Jia, et al. Host-Gut Microbiota Metabolic Interactions. Science. 2012;336(6086):1262-7. [PDF]
- Skillings, D. Holobionts and the ecology of organisms: Multi-species communities or integrated individuals? Biol Philos (2016) 31: 875. [PDF]
- Dyall SD, Brown MT, Johnson PJ. Ancient Invasions: From Endosymbionts to Organelles. Science. 2004;304(5668):253-7. [PDF]
- Redecker D, Kodner R, Graham LE. Glomalean Fungi from the Ordovician. Science. 2000;289(5486):1920 [PDF]
- Heckman DS, Geiser DM, Eidell BR, Stauffer RL, Kardos NL, Hedges SB. Molecular Evidence for the Early Colonization of Land by Fungi and Plants. Science. 2001;293(5532):1129-33. [PDF]
- Muraille E. Redefining the Immune System as a Social Interface for Cooperative Processes. PLoS Pathogens. 2013;9(3). [PDF]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature Biotechnology. 2014;32(4):347-55. [PDF]